INPUTFILETEMPLATE.gjf
1_PESscan.bash
2_bcf.bash
3_gaussian_bcf_top4lines.txt
The above scripts were made to overcome some of the potential energy surface (PES) scan limitations (well, design issues I'd rather not design around) in GAMESS and Gaussian. The intent of the scripts are to:
a) take a molecular input file from some quantum chemical code
b) align the molecule such that the surface you want to perform the PES scan across is aligned in the XY, YZ, or XZ planes
c) define the atom/molecule you want to scan with (such as a cation) in variable form in the input file template
d) define the grid size you want to perform the scan with (how fine a mesh)
e) generate the input files
and finally,
f) write out a command line script in a format that leaves for little post-processing for both the calculation and the analysis.
The demo case will be the (nominally, depending on the level of theory) C2v molecular anion (-1) diphenylmethanide in Gaussian, which will also hit a couple of technical points to speed up the PES scan. The intent was to take a cation (K+, Rb+) and determine the binding energies of the cation/anion pair over the (here, YZ) plane of the molecule to look at preferred binding positions and, more interestingly, the range in binding energies at different positions (see figure below).
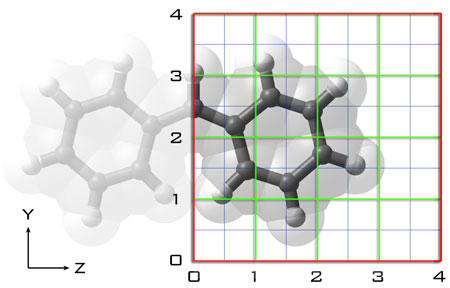
A few setup point for the script (general):
1. The molecule should be aligned along a plane (assuming it is planar. If not, you'll have to tweak the code a little to taste, which isn't too bad) because the script is going to sweep along Y and Z with the assumption that X is a constant vertical value above the plane of the molecule. The goal is to make a 2D map of a surface at one height, then change the height and redo the scan to look at the energy differences in sheets.
2. A molecule optimized in GAMESS and Gaussian should, provided symmetry is employed, orient the molecule such that a molecular plane is aligned along a Cartesian plane. In Gaussian, using the keyword symm(loose,follow) will both nudge a molecule into a higher symmetry and maintain that higher symmetry in the optimization. For the sake of the surface scan, this restriction of the molecule to a higher point group saves significant time, makes the analysis of the script results easier, and will not significantly alter the energies of a molecule whose minimum lies close to a higher symmetry form anyway (the differences in energy between the C2v and Cs diphenylmethanide structures, for instance, is on the order of a few kJ/mol by most levels of theory when the level of theory does NOT predict the C2v form to be the minimum).
3. Negative values are OK in the script, although some shells may not like having the "-" sign in the name of the files. If this is a problem on your machine, take the oriented molecule from Step 1 and add some constant value to all of the X, Y, Z coordinates to make the corner of the scan you start the X,Y,Z analysis from be (X,0,0). I use "X" here because the molecule may best be aligned to the X = 0 plane, which is easiest for some of the later post-processing.
4. If your molecule has two mirror planes (such as diphenylmethanide), set up the scan such that you're only looking at HALF the structure. That should make perfect sense.
The scripts are provided in the links below. Their uses and results are as follows:
Input File 0. INPUTFILETEMPLATE – this file contains the parameters and coordinates for the PES scan. Make sure that this isn't just a cut-and-paste of the optimization file (change the keywords so you run only SINGLE POINT CALCULATIONS!). More on this file below.
Input File 1. 1_PESscan.bash – the bash script that takes a template input file and generates (a) all of the single-point energy input files for the scan and (b) a batch file for submitting all of the files.
Input File 2. 2_bcf.bash – this is Gaussian-specific for the standard Gaussian PC interface, where the script for running multiple jobs is defined in the batch control file (bcf). This script gets the formatting right. Not needed for GAMESS, etc., calculations.
Input File 3. 3_gaussian_bcf_top4lines.txt – this file could be embedded into the 2_bcf.bash file if you like. Part of the 2_bcf.bash script catenates (with cat) the contents of this file and the contents of name files in the right bcf format. This will make perfect sense after the first run.
Output File 4. 4_batchscript.bat – this file (with the numerous input files) is generated from the 1_PESscan.bash script and contains all of the input files and execution parameters for the prompt. Will be Windows- or UNIX-friendly if you did it right.
Output File 5. 5_bcf_for_gaussian.bcf – this is the batch control file for Windows-based Gaussian calculations generated from 2_bcf.bash.
Specifics of each file are provided below:
Input File 0. INPUTFILETEMPLATE
The input template file should look like the following…
%mem=500MB %nproc=2 # scf=tight b3lyp/6-31+g(d,p) integral(grid=ultrafine) diphenylmethanide surface scan 0 1 K REPLACEX REPLACEY REPLACEZ C 0.000000 0.000000 0.940353 C 0.000000 1.308620 0.380671 ...
Note that the molecule is aligned along X=0 and that I replaced opt with scf from the molecular optimization. You don't want to disappear for a weekend only to find that you've done 4 structure optimizations when you could have done 100 single-point calculations. The cation we're sliding with is defined with its X,Y,Z coordinates as REPLACEX,REPLACEY,REPLACEZ. These variables will be replaced by values along the PES grid by running 1_PESscan.bash.
Input File 1. 1_PESscan.bash
The usage of this bash script is:
./1_PESscan.bash $1 $2 $3 $4 $5 $6
or, for instance,
./1_PESscan.bash Kdiphenylmethanide gjf g03 100 25 25
Here are what the variables are.
$1 = name of the input file template (no extension)
$2 = name of the file extension (gjf for Gaussian, inp for GAMESS, etc.)
$3 = command line executable (g03 for Gaussian, gms… for GAMESS
$4 = decimal increments for the X axis (100 = unit, no decimals)
$5 = decimal increments for the Y axis (100 = unit, no decimals)
$6 = decimal increments for the Z axis (100 = unit, no decimals)
You're limited to nine variables in the command line, so there's one modification you need to make to the 1_PESscan.bash file itself. In the first section, MINX through MAXZ need to be defined. These are the integer steps taken by the script to generate the surfaces. These will depend on how you oriented your molecule. The decimal increments will not (which is why they're called in the command line).
Input File 2. 2_bcf.bash
There's nothing to edit here. It will take all of the Gaussian input (.gjf) files in a directory and make the corresponding .bcf file. Major time-saver.
Input File 3. 3_gaussian_bcf_top4lines.txt
This file contains the following…
! !user created batch file list !start=1 !
Which is just the typical 4 top lines in a bcf file (start referring to which molecule in the series gets run first).
And that's the worst of it. Definitely practice the script(s) a few times before beginning a calculation, expect some fuss (depending on how you've pathed your executables) initially, and start with very COARSE grid sizes before wasting a lot of time on generating and checking files (decimal increments of 100 or 50, for instance).
A brief word on the input/output files. The format for the file names generated from 1_PESscan.bash are as follows.
INPUTFILETEMPLATE_X_2.000000_Y_0.000000_Z_0.000000.gjf INPUTFILETEMPLATE_X_2.000000_Y_0.000000_Z_0.500000.gjf INPUTFILETEMPLATE_X_2.000000_Y_0.000000_Z_1.000000.gjf INPUTFILETEMPLATE_X_2.000000_Y_0.000000_Z_1.500000.gjf ...
The format (with six decimal places) is used here so that a user can simply use "ls *.gjf > file.txt" to generate a text file with all of the scanned coordinates, which can then be opened in Excel or something to make the position columns. Further, of course, is that a grep for the final energies in these files will make another file with the coordinates and energies together, which is then easily tweaked and plotted.
That's the worst of it, short of plotting a few surfaces. Any questions or better ideas, drop a line.
